About the TNRS
What is the TNRS?How does the TNRS work?
Where does the TNRS get its taxonomy?
Source code
Contributing software
Contributors
Funding
What is the Taxonomic Name Resolution Service?
The Taxonomic Name Resolution Service (TNRS) is a tool for the computer-assisted standardization of plant scientific names. The TNRS corrects spelling errors and alternative spellings to a standard list of names, and converts out of date names (synonyms) to the current accepted name. The TNRS can process many names at once, saving hours of tedious and error-prone manual name correction. For names that cannot be resolved automatically, the TNRS present a list of possibilities and provides tools for researching and selecting the preferred name.
How does the TNRS work?
The TNRS attempts to match each name submitted to a published scientific name in the TNRS database, correcting spelling if necessary. Once matched, any synonyms are converted to the correct (accepted) name. Both the matched name and the accepted name are returned by the TNRS. This process is performed in the following steps:
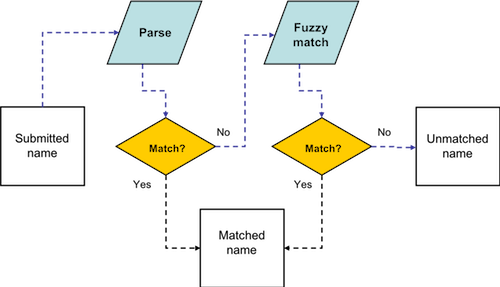
1. Parse. The TNRS first parses (splits) the name into its components parts. Components of a species name include genus, specific epithet, and authority, if included. If the name is a subspecies or variety, the parser will also separate the rank indicator ("var.", "subsp.", "sbsp.", etc.) and the subspecific epithet. The parser also detects and separates standard botanical annotations such as "sp. nov." (new species) and "ined." (unpublished name) as well as indicators of uncertainty such as "cf." ("compare with") and "aff." (affinis, related to but not the same). Finally, any unrecognized components are saved as "Unmatched_Terms". Separating "contaminants" from standard components increases the chance that the TNRS will match the intended name. Parsing is performed by the Global Names Biodiversity Name Parser.
2. Match. The parsed name components are again matched against known scientific names in the TNRS database. The TNRS attempts both exact matching and fuzzy matching using the Taxamatch taxonomic fuzzy matching algorithm. The Taxamatch algorithm speeds up fuzzy matching by searching within the taxonomic hierarchy. For example, once a genus has been identified, only species within that genus are searched.
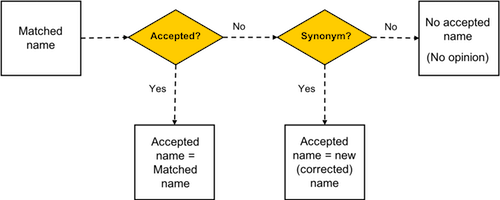
The steps in name matching are illustrated in the figure below:3. Correct. Once the TNRS has discovered the most likely intended scientific name, it will then examine the taxonomic status of that name. If the name is an outdated synonym of another name, the TNRS will return the "Accepted" (correct) name along with matched name, according to the taxonomic sources selected by the user. For some erroneous names, the TNRS will return only the matched name but no accepted name. This can happen is the accepted name is missing or unknown in the selected taxonomic database, or if the name matched is nomenclaturally invalid (e.g., "Invalid", "Illegitimate"), in which case an accepted name may not exist.
The steps in name correction are illustrated in the figure below:3. Select Best Match. Different sources can sometimes return different names as the single correct (accepted) name. Even if you are using only one taxonomic source, a submitted name can sometimes fuzzy match to multiple names with exactly the same match score. In such cases, the TNRS uses a conservative "Best Match Algorithm" to sort the names in descending order of match quality, preferring, for example, synonyms which have been corrected to a different accepted name over the same name labelled as accepted. After applying these rules, the TNRS marks the top-ranked name as the single best match. In such cases, the TNRS will alert you that multiple matches were found, allowing you to select an alternative match if preferred. We recommend that users examine all alternative matches rather than accepting uncritically the TNRS's choice of "Best Match".
Where does the TNRS get its taxonomy?
The TNRS resolves names by consulting one or more publicly-available, expert-curated taxonomic databases. Thus, the TNRS does not provide its own opinions, but simply speeds up the process of researching the status of taxonomic names according to the authoritative sources. Although the TNRS allows users to choose which taxonomic sources they consult, in the end, the opinions provided are those of the selected sources, not the TNRS. For a list of current taxanomic sources, see Sources.
Source code
Source code for all TNRS components is publicly available from the following repositories:
TNRS Search Engine: https://github.com/ojalaquellueva/TNRSbatch
TNRS database: https://github.com/ojalaquellueva/tnrs_db
TNRS name parser: https://github.com/GlobalNamesArchitecture/biodiversity (=Global Names Biodiversity Parser - Ruby version)
TNRS API: https://github.com/ojalaquellueva/TNRSapi
RTNRS R package: https://github.com/EnquistLab/RTNRS
Contributing software
Code from the following open source applications was used during the development of the TNRS:
Original Taxamatch Algorithm. Developed by Tony Rees.
Taxamatch PHP Web Service. Developed by Michael Giddens.
Global Names Biodiversity Parser (Ruby version). Developed by Dmitry Mozzherin.
Contributors
TNRS was first developed by the iPlant Collaborative Tree of Life Project, in collaboration with the Missouri Botanical Garden and the Botanical Information and Ecology Network. Later development was supported by Cyverse, led by the Botanical Information and Ecology Network and funded by a National Science Foundation Harnessing the Data Revolution Grant HDR 1934790. Numerous members of the taxonomic and informatics community provided advice , access to data, and source code.
Project conception and direction
Brad Boyle University of Arizona
Brian Enquist University of Arizona
Application development
Brad Boyle (TNRS database, API and TNRS batch/parallelization update)
Naim Matasci (Original TNRS batch mode/parallelization)
Dmitry Mozzherin (Name parser)
Tony Rees (Fuzzy matching module)
Michael Giddens (Taxamatch PHP adaptation)
George C. Barbosa (TNRSweb Javascript/Node.js user interface)
Rohith Kumar Sajja (TNRSweb Javascript/Node.js user interface)
Rethvick Sriram Yugendra Babu (TNRSweb Javascript/Node.js user interface)
Project direction and development - Original iPlant TNRS (deprecated)
Brad Boyle
Brian Enquist
Juan Antonio Raygoza Garay
Dmitry Mozzherin
Tony Rees
Nicole Hopkins
Zhenyuan Lu
Naim Matasci
Martha Narro
Shannon Oliver
William Piel
Jill Yarmchuk
iPlant staff
Collaborators
Brian Maitner
Cory Merow
Bob Magill (Missouri Botanical Garden)
Chris Freeland (Missouri Botanical Garden)
Chuck Miller (Missouri Botanical Garden)
Peter Jorgensen (Missouri Botanical Garden)
Amy Zanne (University of Missouri, St. Louis)
Peter Stevens (Missouri Botanical Garden)
Jay Paige (Missouri Botanical Garden)
Bob Peet (University of North Carolina at Chapel Hill)
Paul Morris (Harvard University)
Alan Paton (Kew Royal Botanic Gardens and the International Plant Names Index)
Michael Giddens (www.silverbiology.com)
David Remsen (Global Biodiversity Information Facility)
David Patterson (Encyclopedia of Life)
Cam Webb (Harvard University)
Institutions
Funding
Funding provided by the National Science Foundation Plant Cyberinfrastructure Program (grant #DBI-0735191) and National Science Foundation Harnessing the Data Revolution Grant HDR 1934790 to Brian J. Enquist.